New Mutations in Six Genes Found to Increase Alzheimer’s Risk
Researchers have identified new mutations in six genes that may increase the risk of developing Alzheimer’s disease.
Their study involved the genetic analysis of 19 families in Utah with higher-than-normal occurrence of the disease. The scientists found relevant mutations in the genes PELI3, FCHO1, SNAP91, COX6A2, MUC16, and PIDD1.
The analysis also identified rare mutations in three other genes previously associated with increased Alzheimer’s risk — ABCA7, TTR, and NOTCH3.
“Identifying people with increased risk for Alzheimer’s disease before they become symptomatic may lead to earlier and more effective interventions,” Justin Miller, PhD, the study’s co-first author, said in a university press release.
These findings may help pinpoint new potential targets and therapeutic approaches for Alzheimer’s, noted Miller, who is also an assistant professor at the University of Kentucky College of Medicine.
The study, “Analysis of high-risk pedigrees identifies 12 candidate variants for Alzheimer’s disease,” was published in the journal Alzheimer’s & Dementia.
Alzheimer’s is a progressive neurodegenerative disease likely caused by a combination of age-related changes in the brain, along with genetic, environmental, and lifestyle factors.
While common mutations in more than 30 genes have been associated with an increased risk of Alzheimer’s, “it is estimated that more than 40% of the genetic variance of [Alzheimer’s disease] remains uncharacterized,” the researchers wrote.
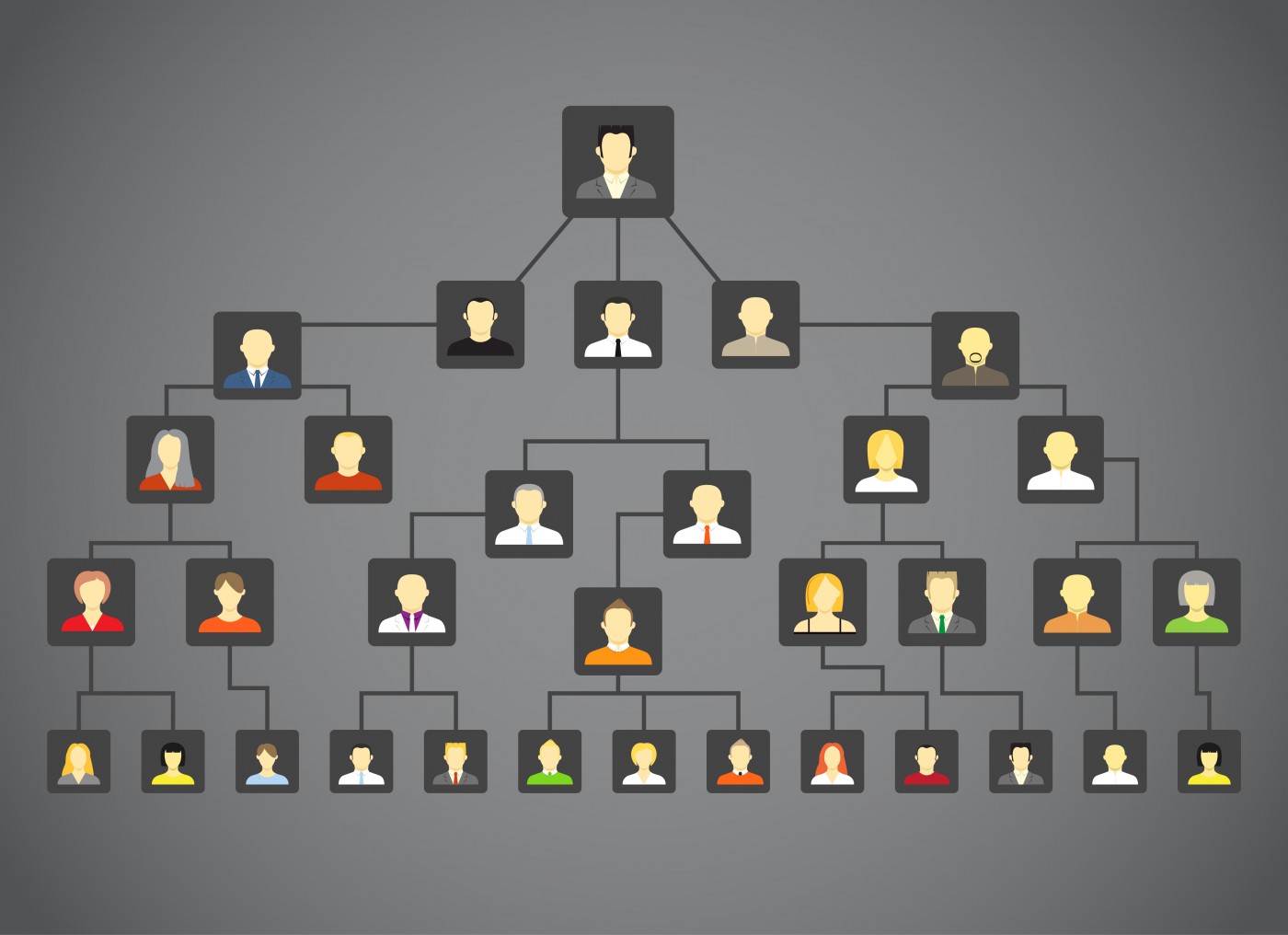
High-risk pedigree studies, or those searching for genetic mutations in families with higher disease occurrence than expected by chance, “are a powerful and efficient method for identification of rare predisposition variants,” the researchers wrote.
This type of study has helped discover genes, such as PLD3, NOTCH3, and RAB10, that contribute to several diseases, including Alzheimer’s.
To identify new genetic risk factors for Alzheimer’s, Miller and his colleagues at Brigham Young University, the University of Utah, and the Alzheimer’s Disease Genetics Consortium, conducted a high-risk pedigree-based analysis using data from the Utah Population Database.
By analyzing the presence of Alzheimer’s-associated death among more than three million people with at least three generations of genealogy in the database, the researchers identified 19 independent families that had a higher-than-normal frequency of the disease and that each included two cousins with Alzheimer’s.
Next, the researchers used a genetic test to examine all protein-coding genes of the 19 cousin pairs and identify mutations shared by both cousins in each family.
“We then used a series of filtering criteria to identify rare genetic variants that were most likely contributing to the excess Alzheimer’s disease in each family,” Miller said.
Mutations shared by at least one cousin pair were compared to publicly available, large-scale datasets related to Alzheimer’s and were considered risk factor candidates if they met any of four criteria.
These criteria included a previous association with Alzheimer’s (known risk variant), an association with increased levels of Alzheimer’s-associated toxic molecules, a higher frequency in a population of Alzheimer’s patients than in an age-matched healthy population, and being present in more than one cousin pair across the 19 families.
The researchers found that 117 mutations in 138 genes shared a biological interaction with genes already implicated in Alzheimer’s. Out of the 117 variants, 11 in nine genes were found to be the best risk factor candidates.
Four mutations in three genes — ABCA7, TTR, and NOTCH3 — had been previously associated with an increased Alzheimer’s risk, further supporting them as risk factors.
Two additional mutations — rs148294193 in the PELI3 gene and rs147599881 in the FCHO1 gene — were associated with increased risk for higher levels of Alzheimer’s-associated toxic forms of amyloid-beta and tau proteins in the cerebrospinal fluid, the liquid that surrounds the brain and spinal cord.
Three genetic variants — rs61729902 in the SNAP91 gene, rs140129800 in the COX6A2 gene, and rs191804178 in the MUC16 gene — were not present in a healthy longevity dataset and were more frequent in Alzheimer’s cases than in healthy controls in another dataset.
Finally, two mutations in the PIDD1 gene (rs200290640 and rs199752248) were present in both members of an Alzheimer’s-affected cousin pair in two independent high-risk families.
While these six genes have not previously been associated with Alzheimer’s, they are known to interact with genes considered potential or confirmed risk factors of the disease.
In addition, identified mutations in the ABCA7, SNAP91, and PELI3 genes were found in other relatives of the original cousin pairs.
“The presence of rare variants identified here may explain the prevalence of [Alzheimer’s disease] mortality in 19 of the 36 [Alzheimer’s]-affected individuals” from these high-risk families, the researchers wrote.
These findings further support ABCA7, TTR, and NOTCH3 as Alzheimer’s risk genes, and identify six other genes as potential risk factors.
They also indicate that “a high-risk pedigree approach can achieve sufficient power to detect rare variants, particularly when coupled with external datasets that contain meaningful data about disease risk,” the researchers wrote.
Among the study’s limitations were the small sample size, the absence of other ethnicities besides white, and the fact that Alzheimer’s diagnoses were based only on death certificate data, which may have missed several other people affected by the disease.
The study was funded by the National Institutes of Health, the Huntsman Cancer Institute, Brigham Young University, the University of Utah, the National Cancer Institute, BrightFocus Foundation, and the National Heart, Lung, and Blood Institute.